First, a prediction algorithm that is based on tabulated chemical shifts for classes of structures , corrected with additive contributions from neighboring functional groups or substructures, is carried out. This method requires first the generation of 3D conformers from a 2D structure so the individual spectra of all conformers are predicted. Next Article Pure shift — experiments in everyday applications — II. Don't have an account? Furthermore, a complementary prediction approach based upon partial atomic charges and steric interactions is also performed. Running Predictions Import a molecular structure. 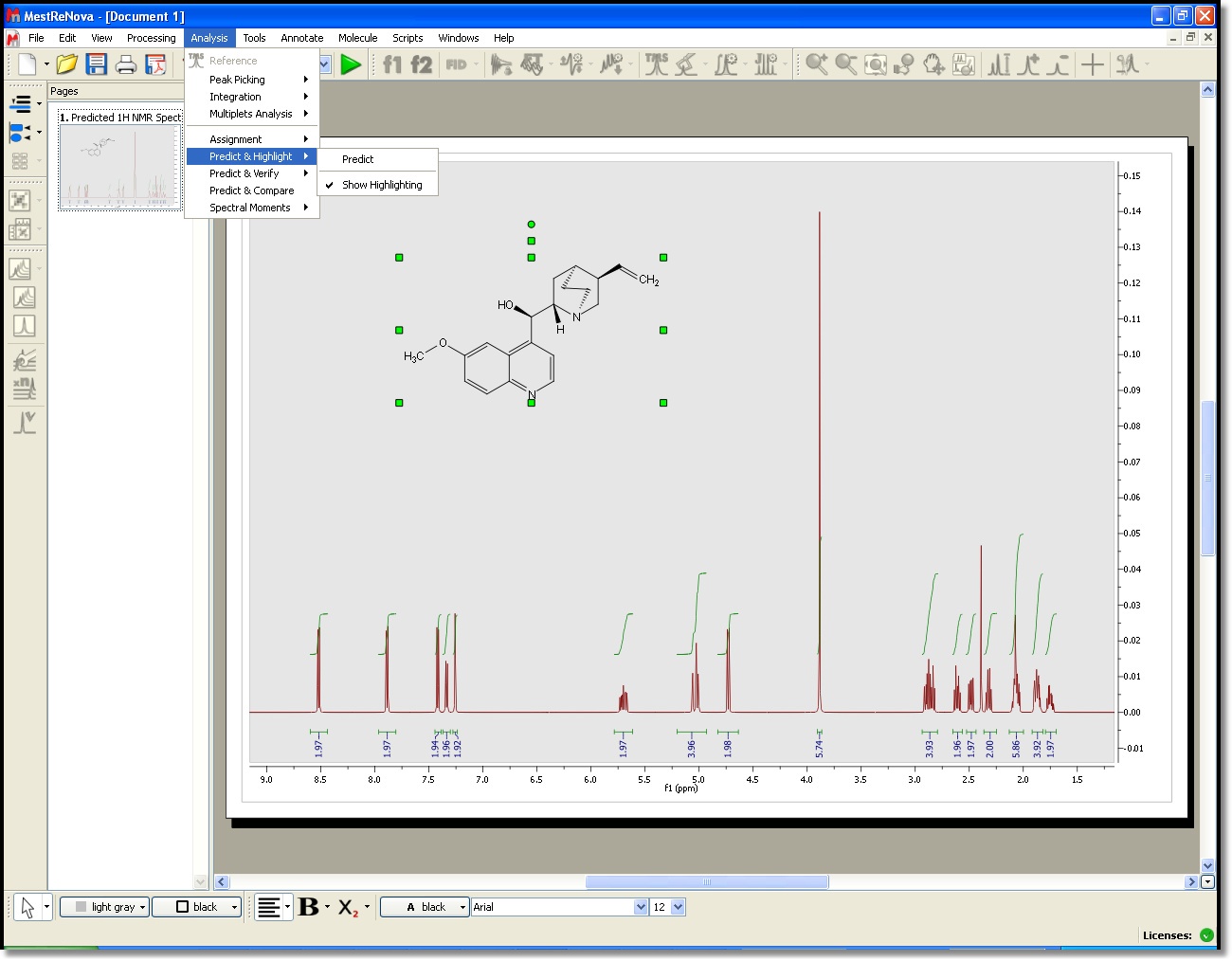
| Uploader: | Kigajas |
| Date Added: | 19 September 2012 |
| File Size: | 45.2 Mb |
| Operating Systems: | Windows NT/2000/XP/2003/2003/7/8/10 MacOS 10/X |
| Downloads: | 1337 |
| Price: | Free* [*Free Regsitration Required] |
Click edit button to change this text. Import a molecular structure.
Starting Guide to NMRPredict Desktop
These substructures provide the base value of a final predicted chemical shift. I am text block. Finally, an average predicted spectrum is calculated employing a Boltzmann weighted desktpp of the shifts calculated for all low-energy conformers. This method requires first the generation of 3D conformers from a 2D structure so the individual spectra of all conformers are predicted. Easily combine and compare experimental and predicted data as part of your workflow.
Ut elit tellus, luctus nec ullamcorper mattis, pulvinar dapibus leo. A similar procedure will be followed to run X-Nuclei predictions: Starting from the atom of interest, all atoms bonded directly to this atom first sphereover two bonds second sphere — and so on — are coded using characters which define atom types, bond types, ring closures, and spheres. This feature will calculate in the background a simulation of the spectrum of the molecular structure present in the spectral window, highlighting the expected chemical shifts when the user hovers the mouse over a proton or a carbon.
Then you will obtain the desired predicted spectrum which can be analyzed as a real one e. In adittion an Mnova Mestrelab Predictor license can be purchased separetely.
Download NMR Predict - Mestrelab
This prediction follows a similar approach to the case of 13C spectra. Don't have an account? Mnova NMR Predict calculates accurate and precise NMR chemical shifts using a novel procedure that combines several prediction engines in a constructive way. The HOSE code approach works very well for query structures that are well represented in the reference collection. Next Article Pure shift — experiments in everyday applications — II. This feature displays, in stacked mode, a simulated spectrum for the molecular structure present in the spectral window, highlighting the expected chemical shifts when the user hovers the mouse over an atom on the molecule.

A password will be e-mailed to you. Currently, molecular structures can be imported as a.
Starting Guide to NMRPredict Desktop - Mestrelab Resources
First, a prediction algorithm that is based on tabulated chemical shifts for classes of structurescorrected with additive contributions from neighboring functional groups or substructures, is carried out. It gives more accurate results when the query atom is not represented in the database.
By Carlos Cobas 9 May, 0. Hovering the mouse over an atom will highlight the area on the spectrum corresponding to the simulated value for that atom.
Previous Article Starting Guide to Mspin. With these two licenses together, you will be entitled to use the new Ensemble Predictor!
Mnova NMR Predict
Lorem ipsum dolor sit amet, consectetur adipiscing elit. This algorithm, named CHARGEis a composite program made up of a neural network based approach for the one- two- and three-bond substituent effects plus a theoretical calculation of the long range effects of substituents.
Prediction of chemical shifts of other nuclides is also available. Running Predictions Import a molecular structure. Furthermore, a complementary prediction approach based upon partial atomic charges and steric interactions is also performed.
The first one is a chemical shift prediction orientated database.

No comments:
Post a Comment